An Intro to R, RStudio, and {tidyverse}
This lab scripts offers what I think to be a gentle introduction to R and RStudio. It will try to acclimate students with R as programming language and RStudio as IDE for the R programming language. There will be a few recurring themes here that are subtle but deceptively critical. 1) You really must know where you are on your computer without referencing to icons you can push (i.e. know your working directory and the path to it). 2) You can push any number of buttons in RStudio, but everything is still a command in a terminal/console. Pay careful attention to that information as it’s communicated to you.
Download the R script for this tutorialElsewhere in My R Cinematic Universe
Some of what I offer here will have to be aggressively plagiarized from other resources I’ve made available. I started teaching graduate-level methods at my previous employer and the bulk of what I wrote there will be ported over here. Likewise, I give basically the same tutorial to our third-semester BA students. There is a much, much older guide that I wrote back in 2014 that you may or may not find super useful. I don’t know what else to say here. When it comes to introducing students to R, you’re going to repeat yourself on loop.
Configure RStudio
When you’re opening R for the very first time, it’ll be useful to just get a general sense of what’s happening. I have a beginner’s guide that I wrote in 2014 (where did the time go!). Notice that I built it around RStudio, which you should download as well. RStudio desktop is free. Don’t pay for a “pro” version. You’re not running a server. You won’t need it.
When you download and install RStudio on top of R, you should customize it just a tiny bit to make the most of the graphical user interface. To do what I recommend doing, select “Tools” in the menu. Scroll to “global options” (which should be at the bottom). On the pop-up, select “pane layout.” Rearrange it so that “Source” is top left, “Console” is top right, and the files/plots/packages/etc. is the bottom right. Thereafter: apply the changes.
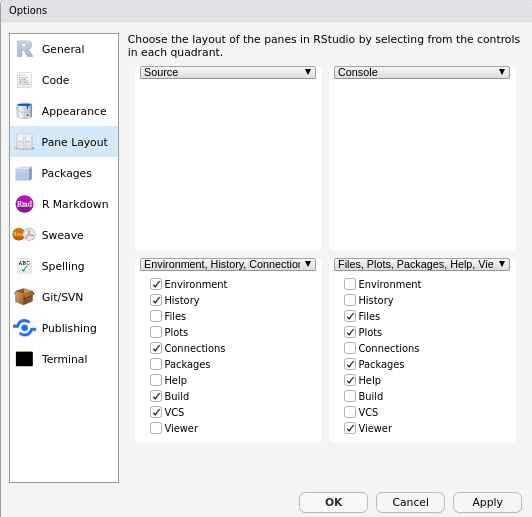
You don’t have to do this, but I think you should since it better economizes space in RStudio. The other pane (environment/history, Git, etc.) is stuff you can either learn to not need (e.g. what’s in the environment) or will only situationally need at an advanced level (e.g. Git information). Minimize that outright. When you’re in RStudio, much of what you’ll be doing leans on the script window and the console window. You’ll occasionally be using the file browser and plot panes as well.
If you have not done so already, open a new script (Ctrl-Shift-N in Windows/Linux or Cmd-Shift-N in Mac) to open a new script.
Get Acclimated in R
Now that you’ve done that, let’s get a general sense of where you are in an R session.
Current Working Directory
First, let’s start with identifying the current working directory. You should know where you are and this happens to be where I am, given the location of this script.
getwd()
#> [1] "/home/steve/Koofr/teaching/eh6127/lab-scripts"
Of note: by default, R’s working directory is the system’s “home” directory. This is somewhat straightforward in Unix-derivative systems, where there is an outright “home” directory. Assume your username is “steve”, then, in Linux, your home directory will be “/home/steve”. In Mac, I think it’s something like “/Users/steve”. Windows users will invariably have something clumsy like “C:/Users/steve/Documents”. Notice the forward slashes. R, like everything else in the world, uses forward slashes. The backslashes owe to Windows’ derivation from DOS.
Create “Objects”
Next, let’s create some “objects.” R is primarily an “object-oriented”
programming language. In as many words, inputs create outputs that may
be assigned to objects in the workspace. You can go nuts here. Of note:
I’ve seen R programmers use =, ->, and <- interchangeably for
object assignment, but I’ve seen instances where = doesn’t work as
intended for object assignment. -> is an option and I use it for
assignment for some complex objects in a “pipe” (more on that later).
For simple cases (and for beginners), lean on <-.
a <- 3
b <- 4
A <- 7
a + b
#> [1] 7
A + b
#> [1] 11
# what objects did we create?
# Notice we did not save a + b or A + b to an object
# Also notice how a pound sign creates a comment? Kinda cool, right?
# Always make comments to yourself.
ls()
#> [1] "a" "A" "b"
Some caution, though. First, don’t create objects with really complex
names. To call them back requires getting every character right in the
console or script. Why inconvenience yourself? Second, R comes with some
default objects that are kinda important and can seriously ruin things
downstream. I don’t know off the top of my head all the default objects
in R, but there are some important ones like TRUE, and FALSE that
you DO NOT want to overwrite. pi is another one you should not
overwrite, and data is a function that serves a specific purpose (even
if you probably won’t be using it a whole lot). You can, however, assign
some built-in objects to new objects.
this_Is_a_long_AND_WEIRD_objEct_name_and_yOu_shoUld_not_do_this <- 5
pi # notice there are a few built-in functions/objects
#> [1] 3.141593
d <- pi # you can assign one built-in object to a new object.
# pi <- 3.14 # don't do this....
If you do something dumb (like overwrite TRUE with something), all
hope is not lost. Just remove the object in question with the rm()
command.
Install/Load Libraries
R depends on user-created libraries to do much of its functionality.
This class will lean on just a few R libraries. The first, {tidyverse}
is our workhorse for workflow. It’ll also be the longest to install
because it comes with lots of dependencies to maximize its
functionality. {stevedata} contains
toy data sets that I use for in-class instruction, and we’ll make use of
these data in these lab sessions (and in your problem sets).
{stevemisc} contains assorted helper
functions that I wrote for my research, which we’ll also use in this
class. {stevetemplates} is not
strictly necessary, but it will make doing your homeworks infinitely
easier (even if you’re not a LaTeX user). {lmtest}, which is not a
package I maintain, does various model diagnostics for OLS.
I may—and probably will, to be honest—ask you to install various other
packages that I think you should have installed. Already, I can see that
the last problem set is going to be a “choose your adventure” at the
end, and request that you have either the {fixest} or {modelr}
package installed. I hope to keep these situations to a minimum.
If any of these result in a “non-zero exit status”, that’s R’s way of
saying “I couldn’t install this.” For you Mac users, the answer to this
situation is probably “update
Xcode.” Xcode is a developer tool
suite for Apple, and many of the {tidyverse} packages require access
to developmental libraries that, to the best of my understanding, are
available in Xcode. In all likelihood, you’re a first-time user who has
not had to think about software development (and so you haven’t updated
Xcode since you first got your Macbook). You might have to do that here.
For you Windows users: I think I’ve figured out what this may look like for you based on my recent foray into the university’s computer labs. The Windows corollary to Xcode is Rtools, which you don’t have installed by default (because it’s not a Microsoft program, per se). You’ll need to install it. First, take inventory of what version of R you have (for the university’s computer labs, it should be 4.0.5). Go to this website and download the version of Rtools that corresponds with the version of R you have. Just click through all the default options so that it can install. Next, in RStudio, open a new blank file and copy-paste the following code into it.
# PATH="${RTOOLS40_HOME}\usr\bin;${PATH}"
I’ll add the caveat that you should remove the hashtag and space preceding that line.
Next, save the file as .Renviron in your default working directory,
which is probably where you are if you are using RStudio for the first
time. The save prompt from RStudio will advise you that this is no
longer an .R file (and, duh, just tell it to save anyway). Afterwards,
restart RStudio and try again. This should fix it, based on my recent
trial run in the university’s computer labs.
For you Linux users: you’re awesome, have great hair, everyone likes you, and you don’t need to worry about a thing, except the various developmental libraries you may have to install from your package repository. My flavor of Linux is in the Debian/Ubuntu family, so here’s an (incomplete) list of developmental libraries based on my experience. Helpfully, most R packages that fail this way will tell you what development library you need, whether in you’re in the Debian or Red Hat family.
If you have yet to install these packages (and you almost certainly have not if you’re opening R for the first time), install it as follows. Note that I’m just commenting out this command so it doesn’t do this when I compile this script on my end.
# Take out the comment...
# install.packages(c("tidyverse", "stevedata", "stevemisc", "stevetemplates", "lmtest"))
Once they’re all installed, you can load the libraries with the
library() command. Of note: you only need to install a package once,
but you’ll need to load the library for each R session. You won’t really
need to load {stevetemplates} for anything since it’s core
functionality is its integration with RStudio. Let’s load {tidyverse}
and {stevedata} in this session, since it’s what I’ll typically use.
library(tidyverse)
#> ── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
#> ✔ dplyr 1.1.4 ✔ readr 2.1.4
#> ✔ forcats 1.0.0 ✔ stringr 1.5.0
#> ✔ ggplot2 4.0.0 ✔ tibble 3.3.0
#> ✔ lubridate 1.9.4 ✔ tidyr 1.3.0
#> ✔ purrr 1.1.0
#> ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ dplyr::lag() masks stats::lag()
#> ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
library(stevedata)
For those of you that are having {tidyverse} installation issues
because of {systemfonts} needing some font-related development
libraries, try this. Again, take out the comments if you want this to
run.
# library(tibble) # special data type we'll use
# library(magrittr) # pipe operator
# library(dplyr) # the workhorse
# library(readr) # for reading particular data types.
# library(stevedata) # for data
These are the core packages that are in {tidyverse} that you should
have installed. Having {tidyverse} loads all of these. It’s basically
a wrapper. Here, you’re just being explicit.
Load Data
Problem sets and lab scripts will lean on data I make available in
{stevedata}. However, you may often find that you want to download a
data set from somewhere else and load it into R. Example data sets would
be stuff like European Values Survey, European Social Survey, or
Varieties of Democracy, or whatever else. You can do this any number of
ways, and it will depend on what is the file format you downloaded. Here
are some commands you’ll want to learn for these circumstances:
haven::read_dta(): for loading Stata .dta fileshaven::read_spss(): for loading SPSS binaries (typically .sav files)read_csv(): for loading comma-separated values (CSV) filesreadxl::read_excel(): for loading MS Excel spreadsheets.read_tsv(): for tab-separated values (TSV) filesreadRDS(): for R serialized data frames, which are awesome for file compression/speed.
Notice that functions like read_dta(), read_spss(), and
read_excel() require some other packages that I didn’t mention.
However, these other packages/libraries are part of the {tidyverse}
and are just not loaded directly with them. Under these conditions, you
can avoid directly loading a library into a session by referencing it
first and grabbing the function you want from within it separated by two
colons (::). Basically, haven::read_dta() could be interpreted as a
command saying “using the {haven} library, grab the read_dta()
command in it”.
These wrappers are also flexible with files on the internet. For example, these will work. Just remember to assign them to an object.
# Note: hypothetical data
Apply <- haven::read_dta("https://stats.idre.ucla.edu/stat/data/ologit.dta")
# Let's take a look at these data.
Apply
#> # A tibble: 400 × 4
#> apply pared public gpa
#> <dbl+lbl> <dbl> <dbl> <dbl>
#> 1 2 [very likely] 0 0 3.26
#> 2 1 [somewhat likely] 1 0 3.21
#> 3 0 [unlikely] 1 1 3.94
#> 4 1 [somewhat likely] 0 0 2.81
#> 5 1 [somewhat likely] 0 0 2.53
#> 6 0 [unlikely] 0 1 2.59
#> 7 1 [somewhat likely] 0 0 2.56
#> 8 1 [somewhat likely] 0 0 2.73
#> 9 0 [unlikely] 0 0 3
#> 10 1 [somewhat likely] 1 0 3.5
#> # ℹ 390 more rows
Learn Some Important R/“Tidy” Functions
I want to spend most of our time in this lab session teaching you some
basic commands you should know to do basically anything in R. These are
so-called “tidy” verbs. We’ll be using some data available in
{stevedata}. This is the pwt_sample data, which includes yearly
economic data for a handful of rich countries that are drawn from
version 10.0 of the Penn World Table. If you’re in RStudio, you can
learn more about these data by typing the following command.
?pwt_sample
I want to dedicate the bulk of this section to learning some core
functions that are part of the {tidyverse}. My introduction here will
inevitably be incomplete because there’s only so much I can teach within
the limited time I have. That said, I’m going to focus on the following
functions available in the {tidyverse} that totally rethink base R.
These are the “pipe” (%>%), glimpse() and summary(), select(),
group_by(), summarize(), mutate(), and filter().
The Pipe (%>%)
I want to start with the pipe because I think of it as the most
important function in the {tidyverse}. The pipe—represented as
%>%—allows you to chain together a series of functions. The pipe is
especially useful if you’re recoding data and you want to make sure you
got everything the way you wanted (and correct) before assigning the
data to another object. You can chain together a lot of {tidyverse}
commands with pipes, but we’ll keep our introduction here rather minimal
because I want to use it to teach about some other things.
glimpse() and summary()
glimpse() and summary() will get you basic descriptions of your
data. Personally, I find summary() more informative than glimpse()
though glimpse() is useful if your data have a lot of variables and
you want to just peek into the data without spamming the R console
without output.
Notice, here, the introduction of the pipe (%>%). In the commands
below, pwt_sample %>% glimpse() is equivalent to
glimpse(pwt_sample), but I like to lean more on pipes than perhaps
others would. My workflow starts with (data) objects, applies various
functions to them, and assigns them to objects. I think you’ll get a lot
of mileage thinking that same way too.
pwt_sample %>% glimpse() # notice the pipe
#> Rows: 1,540
#> Columns: 12
#> $ country <chr> "Australia", "Australia", "Australia", "Australia", "Australia…
#> $ isocode <chr> "AUS", "AUS", "AUS", "AUS", "AUS", "AUS", "AUS", "AUS", "AUS",…
#> $ year <int> 1950, 1951, 1952, 1953, 1954, 1955, 1956, 1957, 1958, 1959, 19…
#> $ pop <dbl> 8.354106, 8.599923, 8.782430, 8.950892, 9.159148, 9.374454, 9.…
#> $ hc <dbl> 2.667302, 2.674344, 2.681403, 2.688482, 2.695580, 2.702696, 2.…
#> $ rgdpna <dbl> 127461.2, 130703.1, 125353.1, 138952.2, 150060.7, 155979.7, 15…
#> $ rgdpo <dbl> 114135.0, 110543.1, 108883.4, 122688.5, 131836.4, 138380.6, 14…
#> $ rgdpe <dbl> 121994.0, 113929.4, 111219.9, 123328.9, 131472.1, 136247.3, 13…
#> $ labsh <dbl> 0.6804925, 0.6804925, 0.6804925, 0.6804925, 0.6804925, 0.68049…
#> $ avh <dbl> 2170.923, 2150.847, 2130.956, 2111.249, 2091.725, 2072.381, 20…
#> $ emp <dbl> 3.429873, 3.523916, 3.591675, 3.653409, 3.731083, 3.811291, 3.…
#> $ rnna <dbl> 639991.2, 690113.6, 704562.4, 733107.3, 771454.2, 810403.7, 83…
pwt_sample %>% summary()
#> country isocode year pop
#> Length:1540 Length:1540 Min. :1950 Min. : 0.1432
#> Class :character Class :character 1st Qu.:1967 1st Qu.: 6.4060
#> Mode :character Mode :character Median :1984 Median : 10.5915
#> Mean :1984 Mean : 35.5251
#> 3rd Qu.:2002 3rd Qu.: 50.7108
#> Max. :2019 Max. :329.0649
#> NA's :2
#> hc rgdpna rgdpo rgdpe
#> Min. :1.242 Min. : 1213 Min. : 1329 Min. : 1234
#> 1st Qu.:2.472 1st Qu.: 151193 1st Qu.: 117905 1st Qu.: 117703
#> Median :2.838 Median : 328992 Median : 291610 Median : 295257
#> Mean :2.814 Mean : 1139533 Mean : 1070787 Mean : 1066654
#> 3rd Qu.:3.202 3rd Qu.: 1103018 3rd Qu.: 939821 3rd Qu.: 933379
#> Max. :3.774 Max. :20563592 Max. :20596346 Max. :20856496
#> NA's :2 NA's :2 NA's :2 NA's :2
#> labsh avh emp rnna
#> Min. :0.3168 Min. :1381 Min. : 0.06547 Min. : 8340
#> 1st Qu.:0.5663 1st Qu.:1679 1st Qu.: 2.91308 1st Qu.: 808493
#> Median :0.6211 Median :1849 Median : 4.56337 Median : 1992211
#> Mean :0.6088 Mean :1857 Mean : 16.13788 Mean : 5439111
#> 3rd Qu.:0.6446 3rd Qu.:2032 3rd Qu.: 20.71151 3rd Qu.: 5306239
#> Max. :0.7709 Max. :2528 Max. :158.29959 Max. :69059064
#> NA's :2 NA's :17 NA's :2 NA's :2
select()
select() is useful for basic (but important) data management. You can
use it to grab (or omit) columns from data. For example, let’s say I
wanted to grab all the columns in the data. I could do that with the
following command.
pwt_sample %>% select(everything()) # grab everything
#> # A tibble: 1,540 × 12
#> country isocode year pop hc rgdpna rgdpo rgdpe labsh avh emp
#> <chr> <chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 Australia AUS 1950 8.35 2.67 127461. 114135. 121994. 0.680 2171. 3.43
#> 2 Australia AUS 1951 8.60 2.67 130703. 110543. 113929. 0.680 2151. 3.52
#> 3 Australia AUS 1952 8.78 2.68 125353. 108883. 111220. 0.680 2131. 3.59
#> 4 Australia AUS 1953 8.95 2.69 138952. 122688. 123329. 0.680 2111. 3.65
#> 5 Australia AUS 1954 9.16 2.70 150061. 131836. 131472. 0.680 2092. 3.73
#> 6 Australia AUS 1955 9.37 2.70 155980. 138381. 136247. 0.680 2072. 3.81
#> 7 Australia AUS 1956 9.60 2.71 156338. 140420. 139239. 0.680 2053. 3.90
#> 8 Australia AUS 1957 9.81 2.72 159762. 141453. 139158. 0.680 2034. 3.95
#> 9 Australia AUS 1958 10.0 2.73 170599. 152677. 148572. 0.680 2015. 3.98
#> 10 Australia AUS 1959 10.2 2.74 181049. 162661. 159401. 0.680 1997. 4.03
#> # ℹ 1,530 more rows
#> # ℹ 1 more variable: rnna <dbl>
Do note this is kind of a redundant command. You could just as well spit the entire data into the console and it would’ve done the same thing. Still, here’s if I wanted everything except wanted to drop the labor share of income variable.
pwt_sample %>% select(-labsh) # grab everything, but drop the labsh variable.
#> # A tibble: 1,540 × 11
#> country isocode year pop hc rgdpna rgdpo rgdpe avh emp rnna
#> <chr> <chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 Australia AUS 1950 8.35 2.67 127461. 114135. 1.22e5 2171. 3.43 6.40e5
#> 2 Australia AUS 1951 8.60 2.67 130703. 110543. 1.14e5 2151. 3.52 6.90e5
#> 3 Australia AUS 1952 8.78 2.68 125353. 108883. 1.11e5 2131. 3.59 7.05e5
#> 4 Australia AUS 1953 8.95 2.69 138952. 122688. 1.23e5 2111. 3.65 7.33e5
#> 5 Australia AUS 1954 9.16 2.70 150061. 131836. 1.31e5 2092. 3.73 7.71e5
#> 6 Australia AUS 1955 9.37 2.70 155980. 138381. 1.36e5 2072. 3.81 8.10e5
#> 7 Australia AUS 1956 9.60 2.71 156338. 140420. 1.39e5 2053. 3.90 8.38e5
#> 8 Australia AUS 1957 9.81 2.72 159762. 141453. 1.39e5 2034. 3.95 8.67e5
#> 9 Australia AUS 1958 10.0 2.73 170599. 152677. 1.49e5 2015. 3.98 9.05e5
#> 10 Australia AUS 1959 10.2 2.74 181049. 162661. 1.59e5 1997. 4.03 9.47e5
#> # ℹ 1,530 more rows
Here’s a more typical case. Assume you’re working with a large data
object and you just want a handful of things. In this case, we have all
these economic data on these 21 countries (ed. we really don’t, but roll
with it), but we just want the GDP data along with the important
identifying information for country and year. Here’s how we’d do that in
the select() function, again with some assistance from the pipe.
pwt_sample %>% select(country, year, rgdpna) # grab just these three columns.
#> # A tibble: 1,540 × 3
#> country year rgdpna
#> <chr> <int> <dbl>
#> 1 Australia 1950 127461.
#> 2 Australia 1951 130703.
#> 3 Australia 1952 125353.
#> 4 Australia 1953 138952.
#> 5 Australia 1954 150061.
#> 6 Australia 1955 155980.
#> 7 Australia 1956 156338.
#> 8 Australia 1957 159762.
#> 9 Australia 1958 170599.
#> 10 Australia 1959 181049.
#> # ℹ 1,530 more rows
Grouping data for grouped functions (group_by(), or .by=)
I think the pipe is probably the most important function in the
{tidyverse} even as a critical reader might note that the pipe is 1) a
port from another package ({magrittr}) and 2) now a part of base R in
a different terminology. Thus, the critical reader (and probably me,
depending on my mood) may note that grouping functions— whether through
group_by() or .by—is probably the most important component of the
{tidyverse}. Basically, group_by() allows you to “split” the data
into various subsets, “apply” various functions to them, and “combine”
them into one output. You might see that terminology
“split-apply-combine” as you learn more about the {tidyverse} and its
development.
Here, let’s do a simple group_by() exercise, while also introducing
you to another function: slice(). We’re going to group by country in
pwt_sample and “slice” the first observation for each group/country.
Notice how we can chain these together with a pipe operator.
# Notice we can chain some pipes together
pwt_sample %>%
# group by country
group_by(country) %>%
# Get me the first observation, by group.
slice(1)
#> # A tibble: 22 × 12
#> # Groups: country [22]
#> country isocode year pop hc rgdpna rgdpo rgdpe labsh avh emp
#> <chr> <chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 Austral… AUS 1950 8.35 2.67 127461. 114135. 121994. 0.680 2171. 3.43
#> 2 Austria AUT 1950 6.98 2.55 56938. 41645. 40430. 0.633 2086. 2.93
#> 3 Belgium BEL 1950 8.63 2.20 85076. 72068. 75250. 0.645 2096. 3.46
#> 4 Canada CAN 1950 13.8 2.48 198071. 176435. 170050. 0.769 2209. 6.12
#> 5 Chile CHL 1950 NA NA NA NA NA NA NA NA
#> 6 Denmark DNK 1950 4.27 2.84 54773. 45143. 44974. 0.642 2050. 1.97
#> 7 Finland FIN 1950 4.01 2.12 29957. 26424. 25824. 0.692 2025. 2.19
#> 8 France FRA 1950 42.5 2.18 372194. 324790. 325674. 0.668 2351. 19.6
#> 9 Germany DEU 1950 68.7 2.43 495069. 358993. 351956. 0.668 2428. 30.9
#> 10 Greece GRC 1950 NA NA NA NA NA NA NA NA
#> # ℹ 12 more rows
#> # ℹ 1 more variable: rnna <dbl>
If you don’t group-by the country first, slice(., 1) will just return
the first observation in the data set.
pwt_sample %>%
# Get me the first observation for each country
slice(1) # womp womp. Forgot to group_by()
#> # A tibble: 1 × 12
#> country isocode year pop hc rgdpna rgdpo rgdpe labsh avh emp
#> <chr> <chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 Australia AUS 1950 8.35 2.67 127461. 114135. 121994. 0.680 2171. 3.43
#> # ℹ 1 more variable: rnna <dbl>
I offer one caveat here. If you’re applying a group-specific function
(that you need just once), it’s generally advisable to “ungroup”
(i.e. ungroup()) as the next function in your pipe chain. As you build
together chains/pipes, the intermediate output you get will advise you
of any “groups” you’ve declared in your data. Don’t lose track of those.
This is incidentally why the {tidyverse} effectively “retired” the
group_by() function for .by as an argument in these functions. .by
will always return un-grouped data whereas group_by() always returns
grouped data.
Observe:
pwt_sample %>%
# group by country
group_by(country) %>%
# Get me the first observation, by group.
slice(1)
#> # A tibble: 22 × 12
#> # Groups: country [22]
#> country isocode year pop hc rgdpna rgdpo rgdpe labsh avh emp
#> <chr> <chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 Austral… AUS 1950 8.35 2.67 127461. 114135. 121994. 0.680 2171. 3.43
#> 2 Austria AUT 1950 6.98 2.55 56938. 41645. 40430. 0.633 2086. 2.93
#> 3 Belgium BEL 1950 8.63 2.20 85076. 72068. 75250. 0.645 2096. 3.46
#> 4 Canada CAN 1950 13.8 2.48 198071. 176435. 170050. 0.769 2209. 6.12
#> 5 Chile CHL 1950 NA NA NA NA NA NA NA NA
#> 6 Denmark DNK 1950 4.27 2.84 54773. 45143. 44974. 0.642 2050. 1.97
#> 7 Finland FIN 1950 4.01 2.12 29957. 26424. 25824. 0.692 2025. 2.19
#> 8 France FRA 1950 42.5 2.18 372194. 324790. 325674. 0.668 2351. 19.6
#> 9 Germany DEU 1950 68.7 2.43 495069. 358993. 351956. 0.668 2428. 30.9
#> 10 Greece GRC 1950 NA NA NA NA NA NA NA NA
#> # ℹ 12 more rows
#> # ℹ 1 more variable: rnna <dbl>
pwt_sample %>%
slice(1, .by=country)
#> # A tibble: 22 × 12
#> country isocode year pop hc rgdpna rgdpo rgdpe labsh avh emp
#> <chr> <chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 Austral… AUS 1950 8.35 2.67 127461. 114135. 121994. 0.680 2171. 3.43
#> 2 Austria AUT 1950 6.98 2.55 56938. 41645. 40430. 0.633 2086. 2.93
#> 3 Belgium BEL 1950 8.63 2.20 85076. 72068. 75250. 0.645 2096. 3.46
#> 4 Canada CAN 1950 13.8 2.48 198071. 176435. 170050. 0.769 2209. 6.12
#> 5 Switzer… CHE 1950 4.61 2.94 114900. 74885. 72672. 0.683 2040. 2.33
#> 6 Chile CHL 1950 NA NA NA NA NA NA NA NA
#> 7 Germany DEU 1950 68.7 2.43 495069. 358993. 351956. 0.668 2428. 30.9
#> 8 Denmark DNK 1950 4.27 2.84 54773. 45143. 44974. 0.642 2050. 1.97
#> 9 Spain ESP 1950 28.1 1.87 144617. 108194. 107746. 0.627 2209. 11.6
#> 10 Finland FIN 1950 4.01 2.12 29957. 26424. 25824. 0.692 2025. 2.19
#> # ℹ 12 more rows
#> # ℹ 1 more variable: rnna <dbl>
summarize()
summarize() creates condensed summaries of your data, for whatever it
is that you want. Here, for example, is a kind of dumb way of seeing how
many observations are in the data. nrow(pwt_sample) works just as
well, but alas…
pwt_sample %>%
# How many observations are in the data?
summarize(n = n())
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 1540
More importantly, summarize() works wonderfully with group_by() or
.by=. For example, for each country (group_by(country)), let’s get
the maximum GDP observed in the data.
pwt_sample %>%
group_by(country) %>%
# Give me the max real GDP observed in the data.
summarize(maxgdp = max(rgdpna, na.rm=T))
#> # A tibble: 22 × 2
#> country maxgdp
#> <chr> <dbl>
#> 1 Australia 1319339.
#> 2 Austria 476657.
#> 3 Belgium 534586.
#> 4 Canada 1873625.
#> 5 Chile 443551.
#> 6 Denmark 311122.
#> 7 Finland 247934.
#> 8 France 2963641.
#> 9 Germany 4312886
#> 10 Greece 378189.
#> # ℹ 12 more rows
.by does the same here.
pwt_sample %>%
# Give me the max real GDP observed in the data, .by country.
summarize(maxgdp = max(rgdpna, na.rm=T), .by=country)
#> # A tibble: 22 × 2
#> country maxgdp
#> <chr> <dbl>
#> 1 Australia 1319339.
#> 2 Austria 476657.
#> 3 Belgium 534586.
#> 4 Canada 1873625.
#> 5 Switzerland 648580.
#> 6 Chile 443551.
#> 7 Germany 4312886
#> 8 Denmark 311122.
#> 9 Spain 1895669
#> 10 Finland 247934.
#> # ℹ 12 more rows
One downside (or feature, depending on your perspective) to
summarize() is that it condenses data and discards stuff that’s not
necessary for creating the condensed output. In the case above, notice
we didn’t ask for what year we observed the maximum GDP for a given
country. We just asked for the maximum. If you wanted something that
would also tell you what year that particular observation was, you’ll
probably want a slice() command in lieu of summarize().
Observe:
pwt_sample %>%
group_by(country) %>%
# translated: give me the row, for each country, in which real GDP is the max (ignoring missing values).
slice(which(rgdpna == max(rgdpna, na.rm=T)))
#> # A tibble: 22 × 12
#> # Groups: country [22]
#> country isocode year pop hc rgdpna rgdpo rgdpe labsh avh emp
#> <chr> <chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 Australia AUS 2018 24.9 3.54 1319339. 1.35e6 1.28e6 0.572 1729. 12.6
#> 2 Austria AUT 2019 8.96 3.38 476657. 4.78e5 4.98e5 0.584 1611. 4.55
#> 3 Belgium BEL 2019 11.5 3.15 534586. 5.18e5 5.89e5 0.595 1586. 4.92
#> 4 Canada CAN 2019 37.4 3.72 1873625. 1.87e6 1.84e6 0.655 1689. 19.3
#> 5 Chile CHL 2019 19.0 3.15 443551. 4.41e5 4.47e5 0.440 1914. 8.10
#> 6 Denmark DNK 2019 5.77 3.60 311122. 3.12e5 3.22e5 0.620 1381. 2.97
#> 7 Finland FIN 2019 5.53 3.50 247934. 2.48e5 2.61e5 0.571 1591. 2.67
#> 8 France FRA 2019 67.4 3.23 2963641. 2.95e6 3.02e6 0.624 1505. 28.5
#> 9 Germany DEU 2019 83.5 3.68 4312886 4.39e6 4.43e6 0.642 1386. 44.8
#> 10 Greece GRC 2007 11.1 2.88 378189. 3.52e5 3.80e5 0.543 2111. 4.86
#> # ℹ 12 more rows
#> # ℹ 1 more variable: rnna <dbl>
# or...
pwt_sample %>%
# translated: give me the row, for each country, in which real GDP is the max (ignoring missing values).
slice(which(rgdpna == max(rgdpna, na.rm=T)), .by=country)
#> # A tibble: 22 × 12
#> country isocode year pop hc rgdpna rgdpo rgdpe labsh avh emp
#> <chr> <chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 Australia AUS 2018 24.9 3.54 1.32e6 1.35e6 1.28e6 0.572 1729. 12.6
#> 2 Austria AUT 2019 8.96 3.38 4.77e5 4.78e5 4.98e5 0.584 1611. 4.55
#> 3 Belgium BEL 2019 11.5 3.15 5.35e5 5.18e5 5.89e5 0.595 1586. 4.92
#> 4 Canada CAN 2019 37.4 3.72 1.87e6 1.87e6 1.84e6 0.655 1689. 19.3
#> 5 Switzerland CHE 2019 8.59 3.70 6.49e5 6.48e5 6.17e5 0.684 1557. 5.01
#> 6 Chile CHL 2019 19.0 3.15 4.44e5 4.41e5 4.47e5 0.440 1914. 8.10
#> 7 Germany DEU 2019 83.5 3.68 4.31e6 4.39e6 4.43e6 0.642 1386. 44.8
#> 8 Denmark DNK 2019 5.77 3.60 3.11e5 3.12e5 3.22e5 0.620 1381. 2.97
#> 9 Spain ESP 2019 46.7 2.99 1.90e6 1.89e6 1.93e6 0.555 1686. 19.9
#> 10 Finland FIN 2019 5.53 3.50 2.48e5 2.48e5 2.61e5 0.571 1591. 2.67
#> # ℹ 12 more rows
#> # ℹ 1 more variable: rnna <dbl>
This is a convoluted way of thinking about summarize(), but you’ll
probably find yourself using it a lot.
mutate()
mutate() is probably the most important {tidyverse} function for
data management/recoding. It will allow you to create new columns while
retaining the original dimensions of the data. Consider it the sister
function to summarize(). But, where summarize() discards, mutate()
retains.
Let’s do something simple with mutate(). For example, the rgdpna
column is real GDP in million 2017 USD. What if we wanted to convert
that million to billions? This is simple with mutate(). Helpfully, you
can create a new column that has both the original/raw data and a
new/recoded variable. This is great for reproducibility in your data
management. One thing I will want to reiterate to you through our
sessions is you should never overwrite raw data you have. Always
create new columns if you’re recoding something.
Anyway, here’s “Wonderw-”… sorry, here’s that new real GDP in billions variable we wanted.
pwt_sample %>%
# Convert rgdpna from real GDP in millions to real GDP in billions
mutate(rgdpnab = rgdpna/1000) %>%
# select just what we want for presentation
select(country:year, rgdpna, rgdpnab)
#> # A tibble: 1,540 × 5
#> country isocode year rgdpna rgdpnab
#> <chr> <chr> <int> <dbl> <dbl>
#> 1 Australia AUS 1950 127461. 127.
#> 2 Australia AUS 1951 130703. 131.
#> 3 Australia AUS 1952 125353. 125.
#> 4 Australia AUS 1953 138952. 139.
#> 5 Australia AUS 1954 150061. 150.
#> 6 Australia AUS 1955 155980. 156.
#> 7 Australia AUS 1956 156338. 156.
#> 8 Australia AUS 1957 159762. 160.
#> 9 Australia AUS 1958 170599. 171.
#> 10 Australia AUS 1959 181049. 181.
#> # ℹ 1,530 more rows
Let’s assume we wanted to create a dummy variable for observations in the data starting from the Great Recession forward? In other words, let’s create a dummy variable for all observations that were in 2008 or later.
pwt_sample %>%
mutate(post_recession = ifelse(year >= 2008, 1, 0)) %>%
select(country:year, post_recession)
#> # A tibble: 1,540 × 4
#> country isocode year post_recession
#> <chr> <chr> <int> <dbl>
#> 1 Australia AUS 1950 0
#> 2 Australia AUS 1951 0
#> 3 Australia AUS 1952 0
#> 4 Australia AUS 1953 0
#> 5 Australia AUS 1954 0
#> 6 Australia AUS 1955 0
#> 7 Australia AUS 1956 0
#> 8 Australia AUS 1957 0
#> 9 Australia AUS 1958 0
#> 10 Australia AUS 1959 0
#> # ℹ 1,530 more rows
Knowing these data go to 2019, we can do this another way as well.
pwt_sample %>%
mutate(post_recession = ifelse(year %in% c(2008:2019), 1, 0)) %>%
select(country:year, post_recession)
#> # A tibble: 1,540 × 4
#> country isocode year post_recession
#> <chr> <chr> <int> <dbl>
#> 1 Australia AUS 1950 0
#> 2 Australia AUS 1951 0
#> 3 Australia AUS 1952 0
#> 4 Australia AUS 1953 0
#> 5 Australia AUS 1954 0
#> 6 Australia AUS 1955 0
#> 7 Australia AUS 1956 0
#> 8 Australia AUS 1957 0
#> 9 Australia AUS 1958 0
#> 10 Australia AUS 1959 0
#> # ℹ 1,530 more rows
Economists typically care about GDP per capita, right? We can create
that kind of data ourselves based on information that we have in
pwt_sample.
pwt_sample %>%
mutate(rgdppc = rgdpna/pop) %>%
select(country:year, rgdpna, pop, rgdppc)
#> # A tibble: 1,540 × 6
#> country isocode year rgdpna pop rgdppc
#> <chr> <chr> <int> <dbl> <dbl> <dbl>
#> 1 Australia AUS 1950 127461. 8.35 15257.
#> 2 Australia AUS 1951 130703. 8.60 15198.
#> 3 Australia AUS 1952 125353. 8.78 14273.
#> 4 Australia AUS 1953 138952. 8.95 15524.
#> 5 Australia AUS 1954 150061. 9.16 16384.
#> 6 Australia AUS 1955 155980. 9.37 16639.
#> 7 Australia AUS 1956 156338. 9.60 16285.
#> 8 Australia AUS 1957 159762. 9.81 16278.
#> 9 Australia AUS 1958 170599. 10.0 17027.
#> 10 Australia AUS 1959 181049. 10.2 17684.
#> # ℹ 1,530 more rows
Notice that mutate() also works beautifully with group_by(). For
example, you may recognize that these data are panel data. We have 21
countries (cross-sectional units) across 70 years (time units). If you
don’t believe me, check this out…
pwt_sample %>%
summarize(n = n(),
min = min(year),
max = max(year),
.by=country) %>%
data.frame
#> country n min max
#> 1 Australia 70 1950 2019
#> 2 Austria 70 1950 2019
#> 3 Belgium 70 1950 2019
#> 4 Canada 70 1950 2019
#> 5 Switzerland 70 1950 2019
#> 6 Chile 70 1950 2019
#> 7 Germany 70 1950 2019
#> 8 Denmark 70 1950 2019
#> 9 Spain 70 1950 2019
#> 10 Finland 70 1950 2019
#> 11 France 70 1950 2019
#> 12 United Kingdom 70 1950 2019
#> 13 Greece 70 1950 2019
#> 14 Ireland 70 1950 2019
#> 15 Iceland 70 1950 2019
#> 16 Italy 70 1950 2019
#> 17 Japan 70 1950 2019
#> 18 Netherlands 70 1950 2019
#> 19 Norway 70 1950 2019
#> 20 Portugal 70 1950 2019
#> 21 Sweden 70 1950 2019
#> 22 United States of America 70 1950 2019
You might know—or should know, as you progress—that some panel methods look for “within” effects inside cross-sectional units by looking at the value of some variable relative to the cross-sectional average for that variable. Let’s use the real GDP per capita variable we can create as an example. Observe what’s going to happen here.
pwt_sample %>%
mutate(rgdppc = rgdpna/pop) %>%
select(country:year, rgdpna, pop, rgdppc) %>%
mutate(meanrgdppc = mean(rgdppc),
diffrgdppc = rgdppc - mean(rgdppc),
.by=country)
#> # A tibble: 1,540 × 8
#> country isocode year rgdpna pop rgdppc meanrgdppc diffrgdppc
#> <chr> <chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 Australia AUS 1950 127461. 8.35 15257. 32112. -16854.
#> 2 Australia AUS 1951 130703. 8.60 15198. 32112. -16914.
#> 3 Australia AUS 1952 125353. 8.78 14273. 32112. -17839.
#> 4 Australia AUS 1953 138952. 8.95 15524. 32112. -16588.
#> 5 Australia AUS 1954 150061. 9.16 16384. 32112. -15728.
#> 6 Australia AUS 1955 155980. 9.37 16639. 32112. -15473.
#> 7 Australia AUS 1956 156338. 9.60 16285. 32112. -15827.
#> 8 Australia AUS 1957 159762. 9.81 16278. 32112. -15834.
#> 9 Australia AUS 1958 170599. 10.0 17027. 32112. -15085.
#> 10 Australia AUS 1959 181049. 10.2 17684. 32112. -14428.
#> # ℹ 1,530 more rows
That diffrgdppc variable practically “centers” the real GDP per capita
variable, and values communicate difference from the mean. This is a
so-called “within” variable, or a transformation of a variable where it
now communicates changes of some variable “within” a cross-sectional
unit.
filter()
filter() is a great diagnostic tool for subsetting your data to look
at particular observations. Notice one little thing, especially if
you’re new to programming. The use of double-equal signs (==) is for
making logical statements where as single-equal signs (=) is for
object assignment or column creation. If you’re using filter(), you’re
probably wanting to find cases where something equals something (==),
is greater than something (>), equal to or greater than something
(>=), is less than something (<), or is less than or equal to
something (<=).
Here, let’s grab just the American observations by filtering to where
isocode == “USA”.
pwt_sample %>%
# give me just the USA observations
filter(isocode == "USA")
#> # A tibble: 70 × 12
#> country isocode year pop hc rgdpna rgdpo rgdpe labsh avh emp
#> <chr> <chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 United Stat… USA 1950 156. 2.58 2.47e6 2.47e6 2.45e6 0.628 1990. 62.8
#> 2 United Stat… USA 1951 158. 2.60 2.67e6 2.66e6 2.62e6 0.634 2032. 65.1
#> 3 United Stat… USA 1952 161. 2.61 2.77e6 2.75e6 2.72e6 0.645 2028. 65.9
#> 4 United Stat… USA 1953 164. 2.62 2.90e6 2.88e6 2.85e6 0.644 2021. 66.8
#> 5 United Stat… USA 1954 167. 2.63 2.89e6 2.87e6 2.84e6 0.637 1998. 65.6
#> 6 United Stat… USA 1955 170. 2.65 3.09e6 3.08e6 3.06e6 0.627 2006. 67.5
#> 7 United Stat… USA 1956 173. 2.66 3.16e6 3.15e6 3.12e6 0.640 1990. 69.1
#> 8 United Stat… USA 1957 176. 2.68 3.23e6 3.22e6 3.19e6 0.639 1963. 69.5
#> 9 United Stat… USA 1958 179. 2.69 3.20e6 3.19e6 3.17e6 0.636 1928. 68.2
#> 10 United Stat… USA 1959 182. 2.71 3.42e6 3.42e6 3.40e6 0.629 1954. 69.8
#> # ℹ 60 more rows
#> # ℹ 1 more variable: rnna <dbl>
We could also use filter() to select observations from the most recent
year.
pwt_sample %>%
# give me the observations from the most recent year.
filter(year == max(year))
#> # A tibble: 22 × 12
#> country isocode year pop hc rgdpna rgdpo rgdpe labsh avh emp
#> <chr> <chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 Australia AUS 2019 25.2 3.55 1.32e6 1.36e6 1.28e6 0.572 1727. 12.9
#> 2 Austria AUT 2019 8.96 3.38 4.77e5 4.78e5 4.98e5 0.584 1611. 4.55
#> 3 Belgium BEL 2019 11.5 3.15 5.35e5 5.18e5 5.89e5 0.595 1586. 4.92
#> 4 Canada CAN 2019 37.4 3.72 1.87e6 1.87e6 1.84e6 0.655 1689. 19.3
#> 5 Switzerland CHE 2019 8.59 3.70 6.49e5 6.48e5 6.17e5 0.684 1557. 5.01
#> 6 Chile CHL 2019 19.0 3.15 4.44e5 4.41e5 4.47e5 0.440 1914. 8.10
#> 7 Germany DEU 2019 83.5 3.68 4.31e6 4.39e6 4.43e6 0.642 1386. 44.8
#> 8 Denmark DNK 2019 5.77 3.60 3.11e5 3.12e5 3.22e5 0.620 1381. 2.97
#> 9 Spain ESP 2019 46.7 2.99 1.90e6 1.89e6 1.93e6 0.555 1686. 19.9
#> 10 Finland FIN 2019 5.53 3.50 2.48e5 2.48e5 2.61e5 0.571 1591. 2.67
#> # ℹ 12 more rows
#> # ℹ 1 more variable: rnna <dbl>
If we do this last part, we’ve converted the panel to a cross-sectional data set.
Don’t Forget to Assign!
When you’re done applying functions/doing whatever to your data, don’t
forget to assign what you’ve done to an object. For simple cases, and
for beginners, I recommend thinking “left-handed” and using <- for
object assignment (as we did above). When you’re doing stuff in the
pipe, my “left-handed” thinking prioritizes the starting data in the
pipe chain. Thus, I tend to use -> for object assignment at the end of
the pipe.
Consider a simple example below. I’m starting with the original data
(pwt_sample). I’m using a simple pipe to create a new variable (within
mutate()) that standardizes the real GDP variable from millions to
billions. Afterward, I’m assigning it to a new object (Data) with
->.
pwt_sample %>%
# convert real GDP to billions
mutate(rgdpnab = rgdpna/1000) -> Data
Data
#> # A tibble: 1,540 × 13
#> country isocode year pop hc rgdpna rgdpo rgdpe labsh avh emp
#> <chr> <chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 Australia AUS 1950 8.35 2.67 127461. 114135. 121994. 0.680 2171. 3.43
#> 2 Australia AUS 1951 8.60 2.67 130703. 110543. 113929. 0.680 2151. 3.52
#> 3 Australia AUS 1952 8.78 2.68 125353. 108883. 111220. 0.680 2131. 3.59
#> 4 Australia AUS 1953 8.95 2.69 138952. 122688. 123329. 0.680 2111. 3.65
#> 5 Australia AUS 1954 9.16 2.70 150061. 131836. 131472. 0.680 2092. 3.73
#> 6 Australia AUS 1955 9.37 2.70 155980. 138381. 136247. 0.680 2072. 3.81
#> 7 Australia AUS 1956 9.60 2.71 156338. 140420. 139239. 0.680 2053. 3.90
#> 8 Australia AUS 1957 9.81 2.72 159762. 141453. 139158. 0.680 2034. 3.95
#> 9 Australia AUS 1958 10.0 2.73 170599. 152677. 148572. 0.680 2015. 3.98
#> 10 Australia AUS 1959 10.2 2.74 181049. 162661. 159401. 0.680 1997. 4.03
#> # ℹ 1,530 more rows
#> # ℹ 2 more variables: rnna <dbl>, rgdpnab <dbl>